Scientists in the Niehrs lab discover a new DNA base modification in mammals
RESEARCH HIGHLIGHTS
07 November – The research group of Prof. Christof Niehrs at the Institute of Molecular Biology (IMB) has discovered a new chemical modification of mammalian DNA called adenine poly-ADP-ribosylation. This discovery adds a surprisingly bulky base modification to the known repertoire of enzyme-catalysed DNA modifications in mammals.
The master instructions of a cell are found in its DNA, which encodes all the information necessary for it to function properly. DNA contains four bases – adenine, thymine, guanine and cytosine (A, T, G and C) – which are like the letters used to write the ‘instructions’. Some of these bases can also be enzymatically modified by the addition of chemical groups, which can act as markers on the DNA to alter gene expression. For example, the base cytosine can be modified into methylcytosine by the addition of a methyl group, which is a marker of transcriptional silence.
ADP-ribosylation is a chemical modification known to occur on proteins, either as a single ADP group (mono-ADP-ribosylation) or a chain of ADP groups (poly-ADP-ribosylation, or PARylation) and is catalysed by the PARP family of ADP-ribose polymerases. However, no one knew whether these modifications occurred in mammalian DNA.
In this study, which was published in Nature Communications, Christof and his group set out for a closer look at this possibility. They first confirmed the presence of PARylation in mouse and human DNA using antibodies against ADP and mass spectrometry, painstakingly excluding all other potential sources of PARylation (such as proteins and RNA). They then proved that reducing expression of the enzyme PARP1 decreased DNA PARylation, while inhibition of another enzyme (PAR glycohydrolases) increased the amount of DNA PARylation. This is important because it argues that DNA PARylation is added or removed from DNA by enzymes in the cell as part of a biological pathway (and not by spontaneous non-enzymatic reactions or DNA damage).
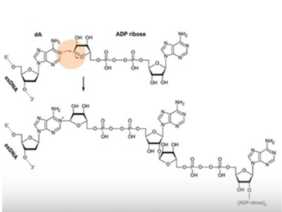
Now that they knew mammalian DNA could be PARylated, the researchers wanted to know where in the genome it occurred. Using in vitro and mass spectrometry studies, they found that PARP1 catalyses PARylation on single-stranded but not double-stranded DNA, and that the modification is added to the base adenine.
The discovery of a previously unknown DNA base modification is extremely important because it could lead to the discovery of new epigenetic processes. “We were excited when we realised we were dealing with an unknown DNA base modification. The major challenge was to pinpoint the identity of this DNA modification down to the chemical linkage between the base and the polymer. We initially couldn’t make sense of the parameters we got from the mass spectrometer. But luckily it finally sunk in”, says Lars Schomacher, co-first and co-corresponding author of the study.
The most important step now is to identify the role of adenine PARylation in the cell. The researchers initially thought that adenine PARylation might be involved in the repair of DNA breaks, but did not find evidence to support this. As the modification only occurs at very low frequencies in mammalian DNA (the researchers calculated there were ~1-2 PARylation sites per genome), the researchers speculate that it might occur transiently in specific circumstances when one strand of DNA is exposed and accessible for PARylation. Lars says, “We are far from concluding that adenine PARylation is an important regulatory mark. Such a bulky base modification seems to be rather incompatible with essential cellular processes like DNA replication or transcription. But because we found it under physiological conditions and it is generated by an important cellular enzyme, we believe it has its function.”
Musheev MU, Schomacher L, Basu A, Han D, Krebs L, Scholz C and Niehrs C (2022) Mammalian N1-adenosine PARylation is a reversible DNA modification. Nat Commun, doi: 10.1038/s41467-022-33731-w Link